SUNDAY NOVEMBER14, 2010
SIMULATING GENE REGULATION IN TH1/2 DIFFERENTIATION
Simulating intracellular gene-regulatory dynamics in agent-based modeling of CD4+ T-cell differentiation
An unbalanced differentiation of T helper cells from precursor type TH0 to the TH1 or TH2 phenotype in immune responses often leads to a pathological condition. In general, immune reactions biased toward TH1 responses may result in auto-immune diseases, while enhanced TH2 responses may cause allergic reactions.
The aim of this work was to integrate a gene regulatory network (GRN) of the TH differentiation in an agent-based model of the hyper-sensitivity reaction. The implementation of such a system introduces a second level of description beyond the mesoscopic level of the inter-cellular interaction of the agent-based model.
The intra-cellular level consists in the cell internal dynamics of gene activation and transcription. The gene regulatory network includes genes-related molecules that have been found to be involved in the differentiation process in TH cells.
The simulator reproduces the hallmarks of an IgE- mediated hypersensitive reaction and provides an example of how to combine the mesoscopic level description of immune cells with the microscopic gene-level dynamics.
Gene network regulating the differentiation of TH cells
The switch of T helper cells from the precursor type TH0 into TH1 and TH2 is a major issue to characterize atopic and non-atopic subjects. In Mendoza (2006) a qualitative model for the regulatory network controlling the TH1/2 differentiation process has been proposed.
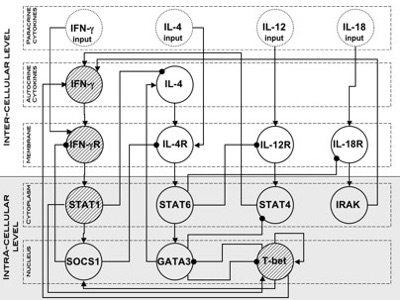
Various kinds of molecules (secreted cytokines, receptors, signal transducers and transcription factors), were considered (see Figure below). Each of the nodes represents a molecule that is known to participate in the differentiation process; the network was built based on published experimental data, all the information comes from in vitro experiments with mouse or human cells (Mendoza (2006)).
The obtained regulatory graph represents the most extensive attempt to model the regulatory network controlling the differentiation of TH lymphocytes to date. The dynamics of the resulting network were analyzed using classical logical method to gain qualitative informations about the dynamical properties of the system (Thomas (1991); Thomas et al. (1995)). In our case, to obtain a dynamics closer to the reality the classical Boolean formalism was extended by allowing the variables to assume three discrete values: low (L), medium (M) and high (H).
The nodes of the gene network represent various kinds of molecules (secreted cytokines, receptors, signal transducers and transcription factors). We consider 17 nodes with some little differences with respect to Mendoza (2006): i) in order to distinguish the input nodes from the internal ones (that have a feedback effect) we added two more nodes, namely, “IFN-γ input” and “IL-4 input”; ii) node IFN-β and IFN-betaR have been omitted since in our model the cytokine IFN-β has overlapping effects with IFN-γ .
Multi-scale simulator
The model at the mesoscopic scale is based on the clonal selection theory of F.M. Burnet (1959) developed on the tracks first highlighted by P. Ehrlich at the beginning of the twenties century.
The present version includes mechanisms related to TH differentiation, isotype switch and histamine release. A cubic millimeter of blood serum is mapped onto a two- dimensional hexagonal lattice (six neighbors), with periodic boundary conditions. Physical proximity is modeled through the concept of lattice-site. All interactions among cells and molecules take place within a lattice-site in a single time step. At the end of each step, entities diffuse on adjacent lattice sites to model cellular and molecular mobility introducing spatial correlations.
Major classes of cells of the lymphoid lineage (lymphocytes T helper and cytotoxic, lymphocytes B and deriving antibody- producing plasma cells) and some of the myeloid lineage (macrophages and mast cells) are represented. Cells are added through an external compartment, which simulate the bone marrow and the thymus. The thymus is implicitly modeled through positive and negative selection of immature thymocytes before they get into the lymphatic system.
The interactions among cells and molecules determine their functional behavior. They may be a-specific (e.g., antigen phagocytosis by monocytes or macrophages, binding by mast cells, etc.) or specific according to their affinity or degree of chemical binding strength (e.g., TH interacting with B cells for antigen recognition, etc.).
At each time step of the simulation all cells and molecules can interact locally (i.e., on each lattice site) according to their internal state, represented by suitable internal variables. An interaction between two cells is considered successful if a change in their internal state has occurred.
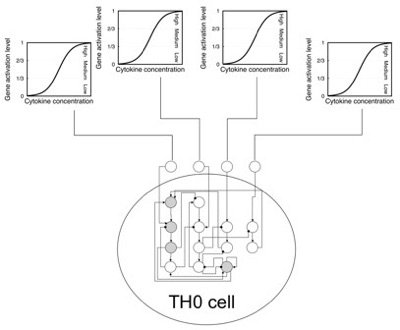
The dynamics of the gene network of each TH cell depends on the concentration of input interleukins surrounding it (i.e., in the same lattice point). These cytokines determine the activation level of the corresponding input nodes (see Figure below).
References
•Mendoza, L. (2006). A network model for the control of the differentiation process in Th cells. Bio Systems, 84(2), 101–114.
This study was conducted under the auspices of the European Community, Project ComplexDIs (contract FP6-2005-NEST-PATH, No.043241).
STUDY